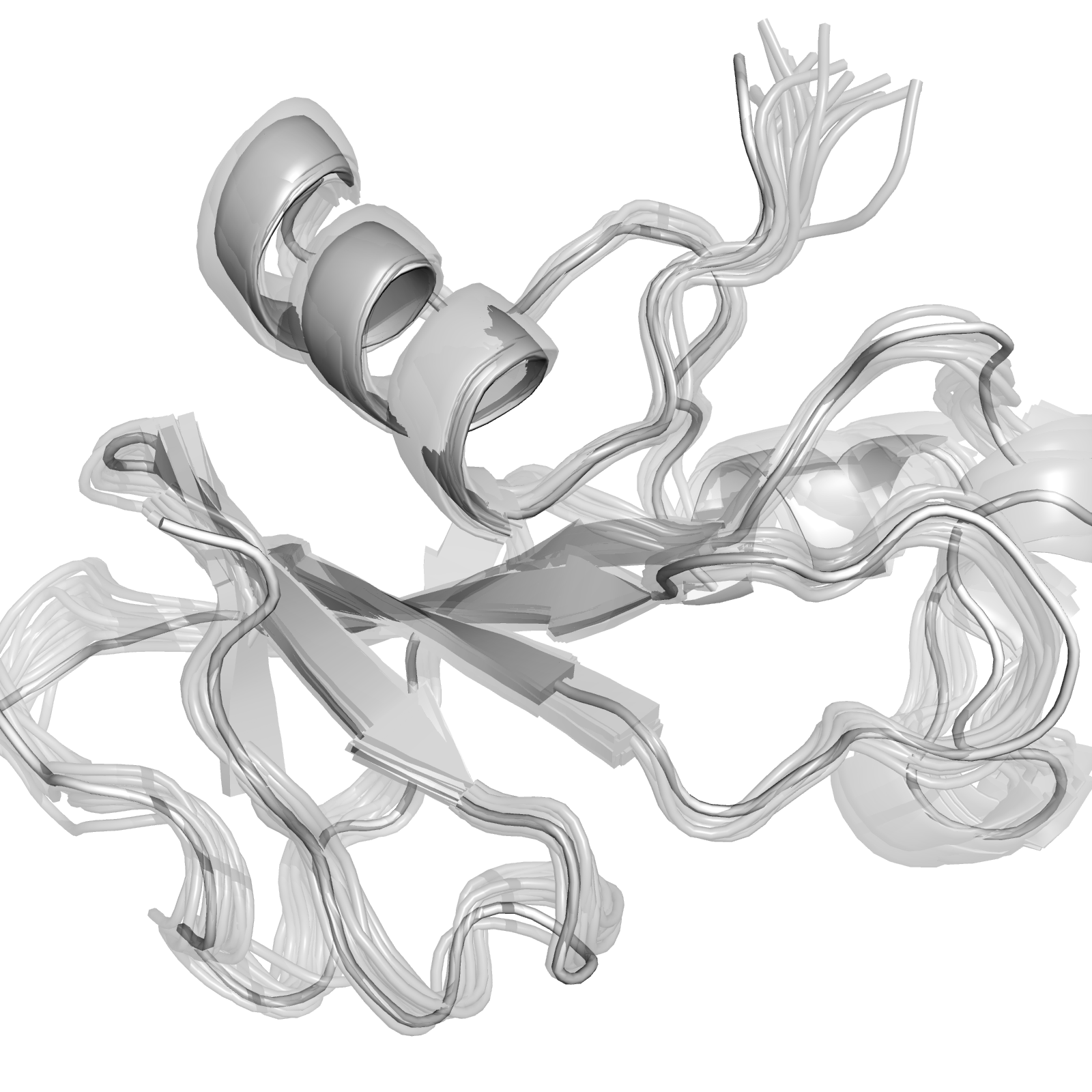
PyMol Trajectory Visualization
Visualize Amber nc trajectory in PyMol
I used to convert everything into .dcd file but it is a little complex. Now I found PyMol actually can directly read prmtop & nc file but just need little adjustment.
Rename .prmtop file
rename .prmtop file into .top file.
REF:
load top file and nc file
load AAA.top |
Note if want to use .mdcrd file from Amber, it needs be renamed as .trj before loading
useful commands in visualization
intra_fit AAA |
https://pymolwiki.org/index.php/Intra_fit
REF:
https://pymolwiki.org/index.php/Load_traj
https://sourceforge.net/p/pymol/mailman/message/36374096/
https://sourceforge.net/p/pymol/mailman/message/35166151/